Opened 3 months ago
Closed 3 months ago
#1613 closed enhancement (fixed)
Include FastQ Screen in the demux step
| Reported by: | Nicklas Nordborg | Owned by: | Nicklas Nordborg |
|---|---|---|---|
| Priority: | major | Milestone: | Reggie v5.3 |
| Component: | net.sf.basedb.reggie | Keywords: | |
| Cc: |
Description (last modified by )
FastQ Screen (https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/) can be used to detect that sequences matches the expected organism.
We should include this as a QC step that is part of the regular demux step and also in the FASTQ import step for RNAseq data. Maybe we can also implement it for the WGS import step.
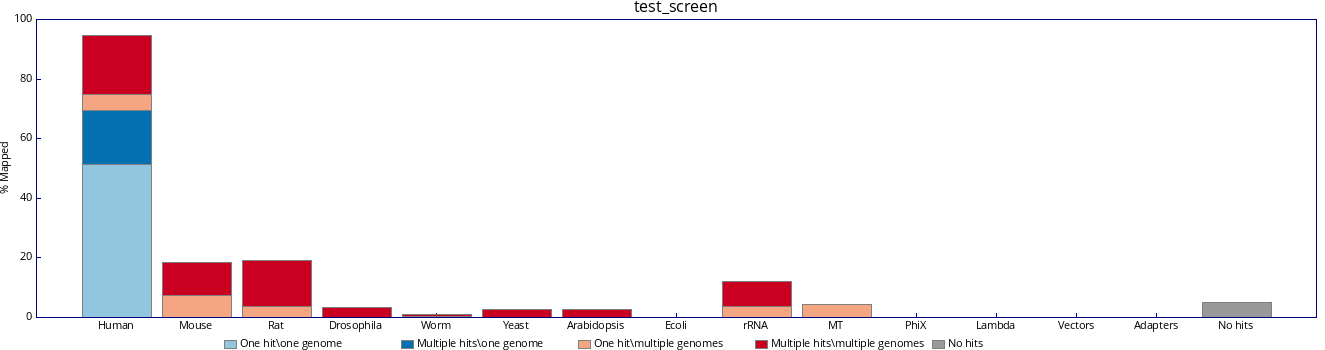
To begin with, we should use the default configuration of databases (see attached image).
We probably want to check the results and issue warnings, etc. Details has not yet been decided.
Attachments (2)
Change History (16)
by , 3 months ago
| Attachment: | test_screen.png added |
|---|
comment:1 by , 3 months ago
| Description: | modified (diff) |
|---|
by , 3 months ago
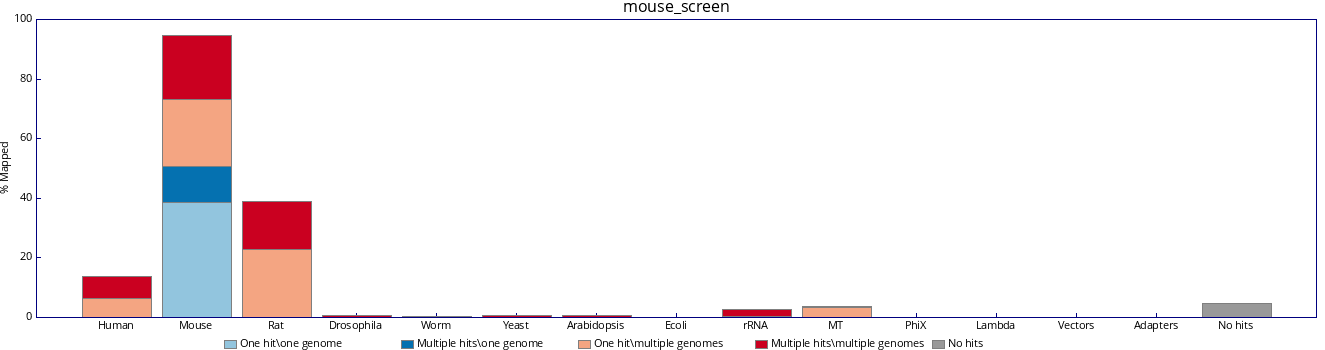
| Attachment: | mouse_screen.png added |
|---|
comment:2 by , 3 months ago
| Description: | modified (diff) |
|---|
comment:3 by , 3 months ago
comment:14 by , 3 months ago
| Resolution: | → fixed |
|---|---|
| Status: | new → closed |
Note:
See TracTickets
for help on using tickets.

In 7854: