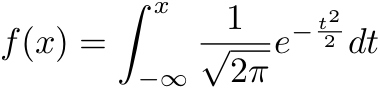| Version 1 (modified by , 17 years ago) ( diff ) |
|---|
One class Z-Test
Uses a z-test to rank genes based on probability for being differentially expressed (DE). Work on bioassays representing two-colour hybridizations equivalent to treated vs. control. Assumes that all the bioassays in the bioassayset are biological replicates.
Calculations
- x is the raw score
- µ is the mean of the population
- σ is the standard deviation of the population.
In statistics, a Z-score is a dimensionless quantity derived by subtracting the population mean from an individual (raw) score and then dividing the difference by the population standard deviation. The z score reveals how many units a case is above or below the mean. The z score allows us to compare the results of different normal distributions, something done with frequent research.
Normsdist returns the probability that the observed value of a standard normal random variable will be less than or equal to z. A standard normal random variable has mean 0 and standard deviation 1 (and also variance 1 because variance = standard deviation squared).
A 2-sided z-test witch corresponds to the probability for a value x where the null hypothesis is that the value is non-differentially expressed (NDE), i.e. a value with low probability is less likely to be NDE.
Population
The population for a specific reporter is represented by all the assays in the bioassayset with a valid M for that reporter. The standard deviation and mean for the population are pooled from the assays. The mean is an average of the means from the assays and the standard deviation is calculated using the equation below.
Equation 4 – Pooled standard deviation
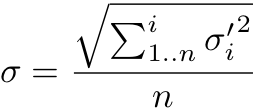
where:
- σ is the pooled standard deviation
- σ’ is the standard deviation from an assay
- n is the number of assays
The plug-in
The plug-in imports a matrix basefile merged on reporter containing reporter and M. For each reporter the plug-in shall calculate:
- The probability, p, based on equation 3 and rank every reporter where rank 1 corresponds to smallest p.
- The number or expected reporters, i.e. the probability * number of reporters.
- The ratio between expected and observed number of reporters, i.e. number of reporters / rank
Output
- A table where each row is a reporter and with column; reporter, M, p, rank, number or expected reporters and ratio between expected and observed.
- Create three plots: “Nmbr vs. Rank”, “nmbrE/Rank vs. Rank”, and a combination of the two.
- Nmbr vs. Rank: A plot with X-values corresponding to Rank. The X-axis is on log scale. Two series of Y-values is plotted; one series with rank (Red line) and one series with number or expected reporters (Blue line).
- nmbrE/Rank vs. Rank: A plot with X-values corresponding to Rank. The X-axis is on log scale. Y-values correspond to ratio between expected and observed.
- A combination plot with two Y-axis containing all three series from plot a and b. Rank (Red line) and number or expected reporters (Blue line) are plotted on one Y-axis and ratio between expected and observed (Black line) is plotted on the other.
closed status tickets:
Attachments (5)
- normsdist.png (6.1 KB ) - added by 17 years ago.
- normsdist.2.png (6.1 KB ) - added by 17 years ago.
- probability.png (3.8 KB ) - added by 17 years ago.
- zscore.png (2.8 KB ) - added by 17 years ago.
- poolSD.png (4.0 KB ) - added by 17 years ago.
Download all attachments as: .zip